kwimage.im_core module¶
Not sure how to best classify these functions
- kwimage.im_core.num_channels(img)[source]¶
Returns the number of color channels in an image.
Assumes images are 2D and the the channels are the trailing dimension. Returns 1 in the case with no trailing channel dimension, otherwise simply returns
img.shape[2].- Parameters:
img (ndarray) – an image with 2 or 3 dimensions.
- Returns:
the number of color channels (1, 3, or 4)
- Return type:
Example
>>> H = W = 3 >>> assert num_channels(np.empty((W, H))) == 1 >>> assert num_channels(np.empty((W, H, 1))) == 1 >>> assert num_channels(np.empty((W, H, 3))) == 3 >>> assert num_channels(np.empty((W, H, 4))) == 4 >>> assert num_channels(np.empty((W, H, 2))) == 2
- kwimage.im_core.ensure_float01(img, dtype=<class 'numpy.float32'>, copy=True)[source]¶
Ensure that an image is encoded using a float32 properly
- Parameters:
img (ndarray) – an image in uint255 or float01 format. Other formats will raise errors.
dtype (type) – a numpy floating type defaults to np.float32
copy (bool) – Always copy if True, else copy if needed. Defaults to True.
- Returns:
an array of floats in the range 0-1
- Return type:
ndarray
- Raises:
ValueError – if the image type is integer and not in [0-255]
Example
>>> ensure_float01(np.array([[0, .5, 1.0]])) array([[0. , 0.5, 1. ]], dtype=float32) >>> ensure_float01(np.array([[0, 1, 200]])) array([[0..., 0.0039..., 0.784...]], dtype=float32)
- kwimage.im_core.ensure_uint255(img, copy=True)[source]¶
Ensure that an image is encoded using a uint8 properly. Either
- Parameters:
img (ndarray) – an image in uint255 or float01 format. Other formats will raise errors.
copy (bool) – always copy if True, else copy if needed. Defaults to True.
- Returns:
an array of bytes in the range 0-255
- Return type:
ndarray
- Raises:
ValueError – if the image type is float and not in [0-1]
ValueError – if the image type is integer and not in [0-255]
Example
>>> ensure_uint255(np.array([[0, .5, 1.0]])) array([[ 0, 127, 255]], dtype=uint8) >>> ensure_uint255(np.array([[0, 1, 200]])) array([[ 0, 1, 200]], dtype=uint8)
- kwimage.im_core.make_channels_comparable(img1, img2, atleast3d=False)[source]¶
Broadcasts image arrays so they can have elementwise operations applied
- Parameters:
img1 (ndarray) – first image
img2 (ndarray) – second image
atleast3d (bool) – if true we ensure that the channel dimension exists (only relevant for 1-channel images). Defaults to False.
Example
>>> import itertools as it >>> wh_basis = [(5, 5), (3, 5), (5, 3), (1, 1), (1, 3), (3, 1)] >>> for w, h in wh_basis: >>> shape_basis = [(w, h), (w, h, 1), (w, h, 3)] >>> # Test all permutations of shap inputs >>> for shape1, shape2 in it.product(shape_basis, shape_basis): >>> print('* input shapes: %r, %r' % (shape1, shape2)) >>> img1 = np.empty(shape1) >>> img2 = np.empty(shape2) >>> img1, img2 = make_channels_comparable(img1, img2) >>> print('... output shapes: %r, %r' % (img1.shape, img2.shape)) >>> elem = (img1 + img2) >>> print('... elem(+) shape: %r' % (elem.shape,)) >>> assert elem.size == img1.size, 'outputs should have same size' >>> assert img1.size == img2.size, 'new imgs should have same size' >>> print('--------')
- kwimage.im_core.atleast_3channels(arr, copy=True)[source]¶
Ensures that there are 3 channels in the image
- Parameters:
arr (ndarray) – an image with 2 or 3 dims.
copy (bool) – Always copies if True, if False, then copies only when the size of the array must change. Defaults to True.
- Returns:
with shape (N, M, C), where C in {3, 4}
- Return type:
ndarray
Doctest
>>> assert atleast_3channels(np.zeros((10, 10))).shape[-1] == 3 >>> assert atleast_3channels(np.zeros((10, 10, 1))).shape[-1] == 3 >>> assert atleast_3channels(np.zeros((10, 10, 3))).shape[-1] == 3 >>> assert atleast_3channels(np.zeros((10, 10, 4))).shape[-1] == 4
- kwimage.im_core.exactly_1channel(image, ndim=2)[source]¶
Returns a 1-channel image as either a 2D or 3D array.
- Parameters:
image (ndarray) – an image with shape (H, W, 1) or (H, W).
ndim (int) – number of dimensions in the output array. Can be either 2 or 3.
- Returns:
if ndim is 2, returns a (H, W) image. if ndim is 3, returns a (H, W, 1) image.
- Return type:
ndarray
- Raises:
ValueError – if assumptions are not met.
Example
>>> import kwimage >>> assert kwimage.exactly_1channel(np.empty((3, 3)), ndim=2).shape == (3, 3) >>> assert kwimage.exactly_1channel(np.empty((3, 3)), ndim=3).shape == (3, 3, 1) >>> assert kwimage.exactly_1channel(np.empty((3, 3, 1)), ndim=2).shape == (3, 3) >>> assert kwimage.exactly_1channel(np.empty((3, 3, 1)), ndim=3).shape == (3, 3, 1) >>> import pytest >>> with pytest.raises(ValueError): >>> kwimage.exactly_1channel(np.empty((3, 3, 2)), ndim=2) >>> with pytest.raises(ValueError): >>> kwimage.exactly_1channel(np.empty((3)), ndim=3)
- kwimage.im_core.padded_slice(data, in_slice, pad=None, padkw=None, return_info=False)[source]¶
Allows slices with out-of-bound coordinates. Any out of bounds coordinate will be sampled via padding.
DEPRECATED FOR THE VERSION IN KWARRAY (slices are more array-ish than image-ish)
Note
Negative slices have a different meaning here then they usually do. Normally, they indicate a wrap-around or a reversed stride, but here they index into out-of-bounds space (which depends on the pad mode). For example a slice of -2:1 literally samples two pixels to the left of the data and one pixel from the data, so you get two padded values and one data value.
- Parameters:
data (Sliceable) – data to slice into. Any channels must be the last dimension.
in_slice (slice | Tuple[slice, …]) – slice for each dimensions
ndim (int) – number of spatial dimensions
pad (List[int|Tuple]) – additional padding of the slice
padkw (Dict) – if unspecified defaults to
{'mode': 'constant'}return_info (bool) – if True, return extra information about the transform. Defaults to False.
- SeeAlso:
_padded_slice_embed - finds the embedded slice and padding _padded_slice_apply - applies padding to sliced data
- Returns:
- data_sliced: subregion of the input data (possibly with padding,
depending on if the original slice went out of bounds)
- Tuple[Sliceable, Dict] :
data_sliced : as above
transform : information on how to return to the original coordinates
- Currently a dict containing:
- st_dims: a list indicating the low and high space-time
coordinate values of the returned data slice.
The structure of this dictionary mach change in the future
- Return type:
Sliceable
Example
>>> data = np.arange(5) >>> in_slice = [slice(-2, 7)]
>>> data_sliced = padded_slice(data, in_slice) >>> print(ub.urepr(data_sliced, with_dtype=False)) np.array([0, 0, 0, 1, 2, 3, 4, 0, 0])
>>> data_sliced = padded_slice(data, in_slice, pad=(3, 3)) >>> print(ub.urepr(data_sliced, with_dtype=False)) np.array([0, 0, 0, 0, 0, 0, 1, 2, 3, 4, 0, 0, 0, 0, 0])
>>> data_sliced = padded_slice(data, slice(3, 4), pad=[(1, 0)]) >>> print(ub.urepr(data_sliced, with_dtype=False)) np.array([2, 3])
- kwimage.im_core._padded_slice_apply(data_clipped, data_slice, extra_padding, padkw=None)[source]¶
Applies requested padding to an extracted data slice.
- kwimage.im_core._padded_slice_embed(in_slice, data_dims, pad=None)[source]¶
Embeds a “padded-slice” inside known data dimension.
Returns the valid data portion of the slice with extra padding for regions outside of the available dimension.
Given a slices for each dimension, image dimensions, and a padding get the corresponding slice from the image and any extra padding needed to achieve the requested window size.
- Parameters:
in_slice (Tuple[slice]) – a tuple of slices for to apply to data data dimension.
data_dims (Tuple[int]) – n-dimension data sizes (e.g. 2d height, width)
pad (tuple) – (List[int|Tuple]): extra pad applied to (left and right) / (both) sides of each slice dim
- Returns:
- data_slice - Tuple[slice] a slice that can be applied to an array
with with shape data_dims. This slice will not correspond to the full window size if the requested slice is out of bounds.
- extra_padding - extra padding needed after slicing to achieve
the requested window size.
- Return type:
Tuple
Example
>>> # Case where slice is inside the data dims on left edge >>> from kwimage.im_core import * # NOQA >>> in_slice = (slice(0, 10), slice(0, 10)) >>> data_dims = [300, 300] >>> pad = [10, 5] >>> a, b = _padded_slice_embed(in_slice, data_dims, pad) >>> print('data_slice = {!r}'.format(a)) >>> print('extra_padding = {!r}'.format(b)) data_slice = (slice(0, 20, None), slice(0, 15, None)) extra_padding = [(10, 0), (5, 0)]
Example
>>> # Case where slice is bigger than the image >>> in_slice = (slice(-10, 400), slice(-10, 400)) >>> data_dims = [300, 300] >>> pad = [10, 5] >>> a, b = _padded_slice_embed(in_slice, data_dims, pad) >>> print('data_slice = {!r}'.format(a)) >>> print('extra_padding = {!r}'.format(b)) data_slice = (slice(0, 300, None), slice(0, 300, None)) extra_padding = [(20, 110), (15, 105)]
Example
>>> # Case where slice is inside than the image >>> in_slice = (slice(10, 40), slice(10, 40)) >>> data_dims = [300, 300] >>> pad = None >>> a, b = _padded_slice_embed(in_slice, data_dims, pad) >>> print('data_slice = {!r}'.format(a)) >>> print('extra_padding = {!r}'.format(b)) data_slice = (slice(10, 40, None), slice(10, 40, None)) extra_padding = [(0, 0), (0, 0)]
- kwimage.im_core.normalize(arr, mode='linear', alpha=None, beta=None, out=None)[source]¶
Rebalance pixel intensities via contrast stretching.
By default linearly stretches pixel intensities to minimum and maximum values.
Note
DEPRECATED: this function has been MOVED to
kwarray.normalize
- kwimage.im_core.find_robust_normalizers(data, params='auto')[source]¶
Finds robust normalization statistics for a single observation
DEPRECATED IN FAVOR of kwarray.find_robust_normalizers
- Parameters:
data (ndarray) – a 1D numpy array where invalid data has already been removed
params (str | dict) – normalization params
- Returns:
normalization parameters
- Return type:
Todo
[ ] No Magic Numbers! Use first principles to deterimine defaults.
[ ] Probably a lot of literature on the subject.
[ ] Is this a kwarray function in general?
Example
>>> from kwimage.im_core import * # NOQA >>> data = np.random.rand(100) >>> norm_params1 = find_robust_normalizers(data, params='auto') >>> norm_params2 = find_robust_normalizers(data, params={'low': 0, 'high': 1.0}) >>> norm_params3 = find_robust_normalizers(np.empty(0), params='auto') >>> print('norm_params1 = {}'.format(ub.urepr(norm_params1, nl=1))) >>> print('norm_params2 = {}'.format(ub.urepr(norm_params2, nl=1))) >>> print('norm_params3 = {}'.format(ub.urepr(norm_params3, nl=1)))
- kwimage.im_core.normalize_intensity(imdata, return_info=False, nodata=None, axis=None, dtype=<class 'numpy.float32'>, params='auto', mask=None)[source]¶
Normalize data intensities using heuristics to help put sensor data with extremely high or low contrast into a visible range.
This function is designed with an emphasis on getting something that is reasonable for visualization.
Todo
[x] Move to kwarray and renamed to robust_normalize?
[ ] Support for M-estimators?
- Parameters:
imdata (ndarray) – raw intensity data
return_info (bool) – if True, return information about the chosen normalization heuristic.
params (str | dict) – can contain keys, low, high, or center e.g. {‘low’: 0.1, ‘center’: 0.8, ‘high’: 0.9}
axis (None | int) – The axis to normalize over, if unspecified, normalize jointly
nodata (None | int) – A value representing nodata to leave unchanged during normalization, for example 0
dtype (type) – can be float32 or float64
mask (ndarray | None) – A mask indicating what pixels are valid and what pixels should be considered nodata. Mutually exclusive with
nodataargument. A mask value of 1 indicates a VALID pixel. A mask value of 0 indicates an INVALID pixel.
- Returns:
a floating point array with values between 0 and 1.
- Return type:
ndarray
Example
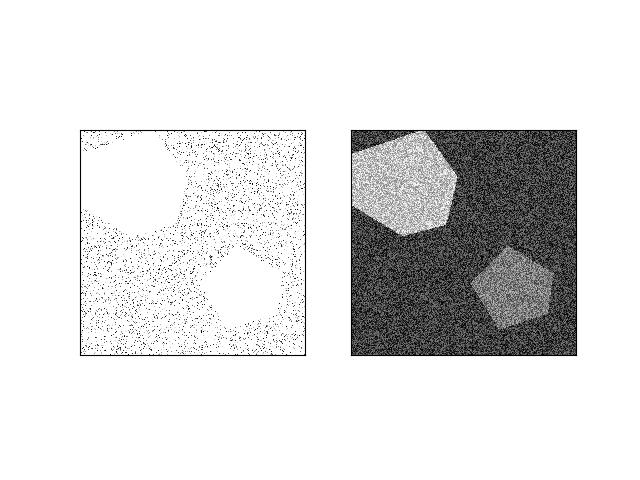
>>> from kwimage.im_core import * # NOQA >>> import ubelt as ub >>> import kwimage >>> import kwarray >>> s = 512 >>> bit_depth = 11 >>> dtype = np.uint16 >>> max_val = int(2 ** bit_depth) >>> min_val = int(0) >>> rng = kwarray.ensure_rng(0) >>> background = np.random.randint(min_val, max_val, size=(s, s), dtype=dtype) >>> poly1 = kwimage.Polygon.random(rng=rng).scale(s / 2) >>> poly2 = kwimage.Polygon.random(rng=rng).scale(s / 2).translate(s / 2) >>> forground = np.zeros_like(background, dtype=np.uint8) >>> forground = poly1.fill(forground, value=255) >>> forground = poly2.fill(forground, value=122) >>> forground = (kwimage.ensure_float01(forground) * max_val).astype(dtype) >>> imdata = background + forground >>> normed, info = normalize_intensity(imdata, return_info=True) >>> print('info = {}'.format(ub.urepr(info, nl=1))) >>> # xdoctest: +REQUIRES(--show) >>> import kwplot >>> kwplot.autompl() >>> kwplot.imshow(imdata, pnum=(1, 2, 1), fnum=1) >>> kwplot.imshow(normed, pnum=(1, 2, 2), fnum=1)
Example
>>> from kwimage.im_core import * # NOQA >>> import ubelt as ub >>> import kwimage >>> # Test on an image that is already normalized to test how it >>> # degrades >>> imdata = kwimage.grab_test_image() / 255
>>> quantile_basis = { >>> 'mode': ['linear', 'sigmoid'], >>> 'high': [0.8, 0.9, 1.0], >>> } >>> quantile_grid = list(ub.named_product(quantile_basis)) >>> quantile_grid += ['auto'] >>> rows = [] >>> rows.append({'key': 'orig', 'result': imdata}) >>> for params in quantile_grid: >>> key = ub.urepr(params, compact=1) >>> result, info = normalize_intensity(imdata, return_info=True, params=params) >>> print('key = {}'.format(key)) >>> print('info = {}'.format(ub.urepr(info, nl=1))) >>> rows.append({'key': key, 'info': info, 'result': result}) >>> # xdoctest: +REQUIRES(--show) >>> import kwplot >>> kwplot.autompl() >>> pnum_ = kwplot.PlotNums(nSubplots=len(rows)) >>> for row in rows: >>> _, ax = kwplot.imshow(row['result'], fnum=1, pnum=pnum_()) >>> ax.set_title(row['key'])
