kwimage.im_cv2 module¶
Wrappers around cv2 functions
Note: all functions in kwimage work with RGB input by default instead of BGR, which is what the underlying cv2 functions use.
- kwimage.im_cv2.connected_components(image, connectivity=8, ltype=<class 'numpy.int32'>, with_stats=True, algo='default')[source]¶
Find connected components in a binary image.
Wrapper around
cv2.connectedComponentsWithStats().- Parameters:
image (ndarray) – a binary uint8 image. Zeros denote the background, and non-zeros numbers are foreground regions that will be partitioned into connected components.
connectivity (int) – either 4 or 8
ltype (numpy.dtype | str | int) – The dtype for the output label array. Can be either ‘int32’ or ‘uint16’, and this can be specified as a cv2 code or a numpy dtype.
algo (str) – The underlying algorithm to use. See [Cv2CCAlgos] for details. Options are spaghetti, sauf, bbdt. (default is spaghetti)
- Returns:
The label array and an information dictionary
- Return type:
Tuple[ndarray, dict]
Todo
Document the details of which type of coordinates we are using. I.e. are pixels points or areas? (I think this uses the points convention?)
Note
opencv 4.5.5 will segfault if connectivity=4 See: [CvIssue21366].
Note
Based on information in [SO35854197].
References
CommandLine
xdoctest -m kwimage.im_cv2 connected_components:0 --show
Example
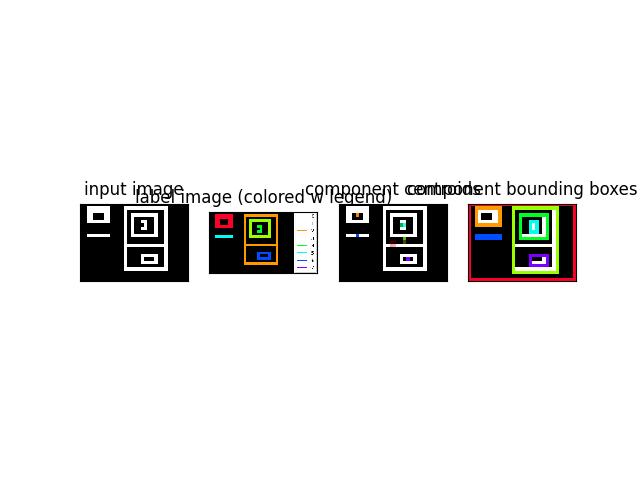
>>> import kwimage >>> from kwimage.im_cv2 import * # NOQA >>> mask = kwimage.Mask.demo() >>> image = mask.data >>> labels, info = connected_components(image) >>> # xdoctest: +REQUIRES(--show) >>> import kwplot >>> kwplot.autompl() >>> canvas0 = kwimage.atleast_3channels(mask.data * 255) >>> canvas2 = canvas0.copy() >>> canvas3 = canvas0.copy() >>> boxes = info['label_boxes'] >>> centroids = info['label_centroids'] >>> label_colors = kwimage.Color.distinct(info['num_labels']) >>> index_to_color = np.array([kwimage.Color('black').as01()] + label_colors) >>> canvas2 = centroids.draw_on(canvas2, color=label_colors, radius=None) >>> boxes.draw_on(canvas3, color=label_colors, thickness=1) >>> legend = kwplot.make_legend_img(ub.dzip(range(len(index_to_color)), index_to_color)) >>> colored_label_img = index_to_color[labels] >>> canvas1 = kwimage.stack_images([colored_label_img, legend], axis=1, resize='smaller') >>> kwplot.imshow(canvas0, pnum=(1, 4, 1), title='input image') >>> kwplot.imshow(canvas1, pnum=(1, 4, 2), title='label image (colored w legend)') >>> kwplot.imshow(canvas2, pnum=(1, 4, 3), title='component centroids') >>> kwplot.imshow(canvas3, pnum=(1, 4, 4), title='component bounding boxes')
- kwimage.im_cv2.convert_colorspace(img, src_space, dst_space, copy=False, implicit=False, dst=None)[source]¶
Converts colorspace of img.
Convenience function around
cv2.cvtColor()- Parameters:
img (ndarray) – image data with float32 or uint8 precision
src_space (str) – input image colorspace. (e.g. BGR, GRAY)
dst_space (str) – desired output colorspace. (e.g. RGB, HSV, LAB)
implicit (bool) –
- if False, the user must correctly specify if the input/output
colorspaces contain alpha channels.
- If True and the input image has an alpha channel, we modify
src_space and dst_space to ensure they both end with “A”.
dst (ndarray[Any, UInt8]) – inplace-output array.
- Returns:
img - image data
- Return type:
ndarray
Note
Note the LAB and HSV colorspaces in float do not go into the 0-1 range.
- For HSV the floating point range is:
0:360, 0:1, 0:1
- For LAB the floating point range is:
0:100, -86.1875:98.234375, -107.859375:94.46875 (Note, that some extreme combinations of a and b are not valid)
Example
>>> import numpy as np >>> convert_colorspace(np.array([[[0, 0, 1]]], dtype=np.float32), 'RGB', 'LAB') >>> convert_colorspace(np.array([[[0, 1, 0]]], dtype=np.float32), 'RGB', 'LAB') >>> convert_colorspace(np.array([[[1, 0, 0]]], dtype=np.float32), 'RGB', 'LAB') >>> convert_colorspace(np.array([[[1, 1, 1]]], dtype=np.float32), 'RGB', 'LAB') >>> convert_colorspace(np.array([[[0, 0, 1]]], dtype=np.float32), 'RGB', 'HSV')
- kwimage.im_cv2.gaussian_blur(image, kernel=None, sigma=None, border_mode=None, dst=None)[source]¶
Apply a gausian blur to an image.
This is a simple wrapper around
cv2.GaussianBlur()with concise parametarization and sane defaults.- Parameters:
image (ndarray) – the input image
kernel (int | Tuple[int, int]) – The kernel size in x and y directions.
sigma (float | Tuple[float, float]) – The gaussian spread in x and y directions.
border_mode (str | int | None) – Border text code or cv2 integer. Border codes are ‘constant’ (default), ‘replicate’, ‘reflect’, ‘reflect101’, and ‘transparent’.
dst (ndarray | None) – optional inplace-output array.
- Returns:
the blurred image
- Return type:
ndarray
Example
>>> import kwimage >>> image = kwimage.ensure_float01(kwimage.grab_test_image('astro')) >>> blurred1 = kwimage.gaussian_blur(image) >>> blurred2 = kwimage.gaussian_blur(image, kernel=9) >>> blurred3 = kwimage.gaussian_blur(image, sigma=2) >>> blurred4 = kwimage.gaussian_blur(image, sigma=(2, 5), kernel=5) >>> # xdoctest: +REQUIRES(--show) >>> import kwplot >>> kwplot.autompl() >>> pnum_ = kwplot.PlotNums(nRows=4, nCols=1) >>> blurs = [blurred1, blurred2, blurred3, blurred4] >>> for blurred in blurs: >>> diff = np.abs(image - blurred) >>> stack = kwimage.stack_images([image, blurred, diff], pad=10, axis=1) >>> kwplot.imshow(stack, pnum=pnum_()) >>> kwplot.show_if_requested()

- kwimage.im_cv2.gaussian_patch(shape=(7, 7), sigma=None)[source]¶
Creates a 2D gaussian patch with a specific size and sigma
- Parameters:
shape (Tuple[int, int]) – patch height and width
sigma (float | Tuple[float, float] | None) – Gaussian standard deviation. If unspecified, it is derived using the formula
0.3 * ((s - 1) * 0.5 - 1) + 0.8as described in [Cv2GaussKern].
- Returns:
ndarray
References
Todo
[ ] Look into this C-implementation https://kwgitlab.kitware.com/computer-vision/heatmap/blob/master/heatmap/heatmap.c
CommandLine
xdoctest -m kwimage.im_cv2 gaussian_patch --show
Example
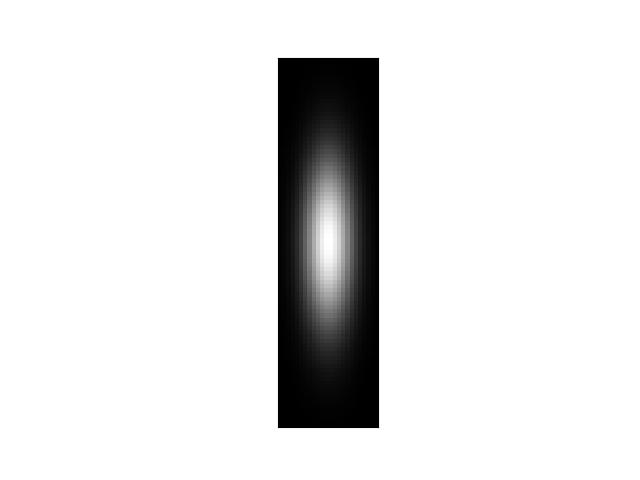
>>> import numpy as np >>> shape = (88, 24) >>> sigma = None # 1.0 >>> gausspatch = gaussian_patch(shape, sigma) >>> sum_ = gausspatch.sum() >>> assert np.all(np.isclose(sum_, 1.0)) >>> # xdoctest: +REQUIRES(--show) >>> import kwplot >>> kwplot.autompl() >>> norm = (gausspatch - gausspatch.min()) / (gausspatch.max() - gausspatch.min()) >>> kwplot.imshow(norm) >>> kwplot.show_if_requested()
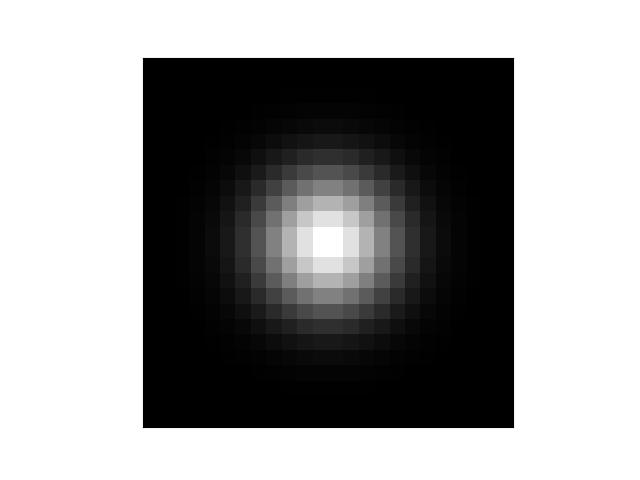
Example
>>> import numpy as np >>> shape = (24, 24) >>> sigma = 3.0 >>> gausspatch = gaussian_patch(shape, sigma) >>> sum_ = gausspatch.sum() >>> assert np.all(np.isclose(sum_, 1.0)) >>> # xdoctest: +REQUIRES(--show) >>> import kwplot >>> kwplot.autompl() >>> norm = (gausspatch - gausspatch.min()) / (gausspatch.max() - gausspatch.min()) >>> kwplot.imshow(norm) >>> kwplot.show_if_requested()
- kwimage.im_cv2.imcrop(img, dsize, about=None, origin=None, border_value=None, interpolation='nearest')[source]¶
Crop an image about a specified point, padding if necessary.
This is like
PIL.Image.Image.crop()with more convenient arguments, orcv2.getRectSubPix()without the baked-in bilinear interpolation.- Parameters:
img (ndarray) – image to crop
dsize (Tuple[None | int, None | int]) – the desired width and height of the new image. If a dimension is None, then it is automatically computed to preserve aspect ratio. This can be larger than the original dims; if so, the cropped image is padded with border_value.
about (Tuple[str | int, str | int]) – the location to crop about. Mutually exclusive with origin. Defaults to top left. If ints (w,h) are provided, that will be the center of the cropped image. There are also string codes available: ‘lt’: make the top left point of the image the top left point of the cropped image. This is equivalent to
img[:dsize[1], :dsize[0]], plus padding. ‘rb’: make the bottom right point of the image the bottom right point of the cropped image. This is equivalent toimg[-dsize[1]:, -dsize[0]:], plus padding. ‘cc’: make the center of the image the center of the cropped image. Any combination of these codes can be used, ex. ‘lb’, ‘ct’, (‘r’, 200), …origin (Tuple[int, int] | None) – the origin of the crop in (x,y) order (same order as dsize/about). Mutually exclusive with about. Defaults to top left.
border_value (Number | Tuple | str) – any border border_value accepted by cv2.copyMakeBorder, ex. [255, 0, 0] (blue). Default is 0.
interpolation (str) – Can be ‘nearest’, in which case integral cropping is used. Can also be ‘linear’, in which case cv2.getRectSubPix is used. Defaults to ‘nearest’.
- Returns:
the cropped image
- Return type:
ndarray
- SeeAlso:
kwarray.padded_slice()- a similar function for working with“negative slices”.
Example
>>> import kwimage >>> import numpy as np >>> # >>> img = kwimage.grab_test_image('astro', dsize=(32, 32))[..., 0:3] >>> # >>> # regular crop >>> new_img1 = kwimage.imcrop(img, dsize=(5,6)) >>> assert new_img1.shape[0:2] == (6, 5) >>> # >>> # padding for coords outside the image bounds >>> new_img2 = kwimage.imcrop(img, dsize=(5,6), >>> origin=(-1,0), border_value=[1, 0, 0]) >>> assert np.all(new_img2[:, 0, 0:3] == [1, 0, 0]) >>> # >>> # codes for corner- and edge-centered cropping >>> new_img3 = kwimage.imcrop(img, dsize=(5,6), >>> about='cb') >>> # >>> # special code for bilinear interpolation >>> # with floating-point coordinates >>> new_img4 = kwimage.imcrop(img, dsize=(5,6), >>> about=(5.5, 8.5), interpolation='linear') >>> # >>> # use with bounding boxes >>> bbox = kwimage.Boxes.random(scale=5, rng=132).to_xywh().quantize() >>> origin, dsize = np.split(bbox.data[0], 2) >>> new_img5 = kwimage.imcrop(img, dsize=dsize, >>> origin=origin) >>> # xdoctest: +REQUIRES(--show) >>> import kwplot >>> kwplot.autompl() >>> pnum_ = kwplot.PlotNums(nSubplots=6) >>> kwplot.imshow(img, pnum=pnum_()) >>> kwplot.imshow(new_img1, pnum=pnum_()) >>> kwplot.imshow(new_img2, pnum=pnum_()) >>> kwplot.imshow(new_img3, pnum=pnum_()) >>> kwplot.imshow(new_img4, pnum=pnum_()) >>> kwplot.imshow(new_img5, pnum=pnum_()) >>> kwplot.show_if_requested()

- kwimage.im_cv2.imresize(img, scale=None, dsize=None, max_dim=None, min_dim=None, interpolation=None, grow_interpolation=None, letterbox=False, return_info=False, antialias=False, border_value=0)[source]¶
Resize an image via a scale factor, final size, or size and aspect ratio.
Wraps and generalizes cv2.resize, allows for specification of either a scale factor, a final size, or the final size for a particular dimension.
Note
As described in [ResizeConfusion], this each entry in the image array as representing the center of a pixel. This is the pixels_are=’area’ approach, or align_corners=False in pytorch. It is equivalent to a shift and scale in warp_affine (which by default uses align corners).
Note
The border mode cannot be specified here and seems to always be reflect in the underlying cv2 implementation.
- Parameters:
img (ndarray) – image to resize
scale (float | Tuple[float, float]) – Desired floating point scale factor. If a tuple, the dimension ordering is x,y. Mutually exclusive with dsize, min_dim, max_dim.
dsize (Tuple[int | None, int | None] | None) – The desired width and height of the new image. If a dimension is None, then it is automatically computed to preserve aspect ratio. Mutually exclusive with scale, min_dim, max_dim.
max_dim (int) – New size of the maximum dimension, the other dimension is scaled to maintain aspect ratio. Mutually exclusive with scale, dsize, min_dim.
min_dim (int) – New size of the minimum dimension, the other dimension is scaled to maintain aspect ratio. Mutually exclusive with scale, dsize, max_dim.
interpolation (str | int) – The interpolation key or code (e.g. linear lanczos). By default “area” is used if the image is shrinking and “lanczos” is used if the image is growing. Note, if this is explicitly set, then it will be used regardless of if the image is growing or shrinking. Set
grow_interpolationto change the default for an enlarging interpolation.grow_interpolation (str | int) – The interpolation key or code to use when the image is being enlarged. Does nothing if “interpolation” is explicitly given. If “interpolation” is not specified “area” is used when shrinking. Defaults to “lanczos”.
letterbox (bool) – If used in conjunction with dsize, then the image is scaled and translated to fit in the center of the new image while maintaining aspect ratio. Border padding is added if necessary. Defaults to False.
return_info (bool) – if True returns information about the final transformation in a dictionary. If there is an offset, the scale is applied before the offset when transforming to the new resized space. Defaults to False.
antialias (bool) – if True blurs to anti-alias before downsampling. Defaults to False.
border_value (int | float | Iterable[int | float]) – if letterbox is True, this is used as the constant fill value.
- Returns:
the new image and optionally an info dictionary if return_info=True
- Return type:
ndarray | Tuple[ndarray, Dict]
References
Example
>>> import kwimage >>> import numpy as np >>> # Test scale >>> img = np.zeros((16, 10, 3), dtype=np.uint8) >>> new_img, info = kwimage.imresize(img, scale=.85, >>> interpolation='area', >>> return_info=True) >>> print('info = {!r}'.format(info)) >>> assert info['scale'].tolist() == [.8, 0.875] >>> # Test dsize without None >>> new_img, info = kwimage.imresize(img, dsize=(5, 12), >>> interpolation='area', >>> return_info=True) >>> print('info = {!r}'.format(info)) >>> assert info['scale'].tolist() == [0.5 , 0.75] >>> # Test dsize with None >>> new_img, info = kwimage.imresize(img, dsize=(6, None), >>> interpolation='area', >>> return_info=True) >>> print('info = {!r}'.format(info)) >>> assert info['scale'].tolist() == [0.6, 0.625] >>> # Test max_dim >>> new_img, info = kwimage.imresize(img, max_dim=6, >>> interpolation='area', >>> return_info=True) >>> print('info = {!r}'.format(info)) >>> assert info['scale'].tolist() == [0.4 , 0.375] >>> # Test min_dim >>> new_img, info = kwimage.imresize(img, min_dim=6, >>> interpolation='area', >>> return_info=True) >>> print('info = {!r}'.format(info)) >>> assert info['scale'].tolist() == [0.6 , 0.625]
Example
>>> import kwimage >>> import numpy as np >>> # Test letterbox resize >>> img = np.ones((5, 10, 3), dtype=np.float32) >>> new_img, info = kwimage.imresize(img, dsize=(19, 19), >>> letterbox=True, >>> return_info=True) >>> print('info = {!r}'.format(info)) >>> assert info['offset'].tolist() == [0, 4] >>> img = np.ones((10, 5, 3), dtype=np.float32) >>> new_img, info = kwimage.imresize(img, dsize=(19, 19), >>> letterbox=True, >>> return_info=True) >>> print('info = {!r}'.format(info)) >>> assert info['offset'].tolist() == [4, 0]
>>> import kwimage >>> import numpy as np >>> # Test letterbox resize >>> img = np.random.rand(100, 200) >>> new_img, info = kwimage.imresize(img, dsize=(300, 300), letterbox=True, return_info=True)
Example
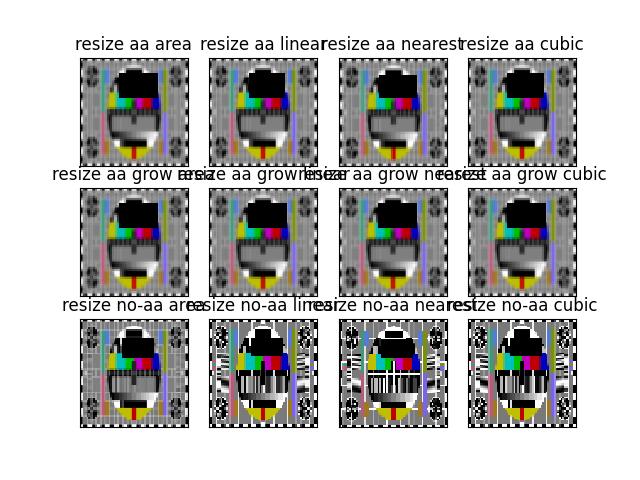
>>> # Check aliasing >>> import kwimage >>> #img = kwimage.grab_test_image('checkerboard') >>> img = kwimage.grab_test_image('pm5644') >>> # test with nans >>> img = kwimage.ensure_float01(img) >>> img[100:200, 400:700] = np.nan >>> # xdoctest: +REQUIRES(--show) >>> import kwplot >>> kwplot.autompl() >>> dsize = (14, 14) >>> dsize = (64, 64) >>> # When we set "grow_interpolation" for a "shrinking" resize it should >>> # still do the "area" interpolation to antialias the results. But if we >>> # use explicit interpolation it should alias. >>> pnum_ = kwplot.PlotNums(nSubplots=12, nCols=4) >>> kwplot.imshow(kwimage.imresize(img, dsize=dsize, antialias=True, interpolation='area'), pnum=pnum_(), title='resize aa area') >>> kwplot.imshow(kwimage.imresize(img, dsize=dsize, antialias=True, interpolation='linear'), pnum=pnum_(), title='resize aa linear') >>> kwplot.imshow(kwimage.imresize(img, dsize=dsize, antialias=True, interpolation='nearest'), pnum=pnum_(), title='resize aa nearest') >>> kwplot.imshow(kwimage.imresize(img, dsize=dsize, antialias=True, interpolation='cubic'), pnum=pnum_(), title='resize aa cubic')
>>> kwplot.imshow(kwimage.imresize(img, dsize=dsize, antialias=True, grow_interpolation='area'), pnum=pnum_(), title='resize aa grow area') >>> kwplot.imshow(kwimage.imresize(img, dsize=dsize, antialias=True, grow_interpolation='linear'), pnum=pnum_(), title='resize aa grow linear') >>> kwplot.imshow(kwimage.imresize(img, dsize=dsize, antialias=True, grow_interpolation='nearest'), pnum=pnum_(), title='resize aa grow nearest') >>> kwplot.imshow(kwimage.imresize(img, dsize=dsize, antialias=True, grow_interpolation='cubic'), pnum=pnum_(), title='resize aa grow cubic')
>>> kwplot.imshow(kwimage.imresize(img, dsize=dsize, antialias=False, interpolation='area'), pnum=pnum_(), title='resize no-aa area') >>> kwplot.imshow(kwimage.imresize(img, dsize=dsize, antialias=False, interpolation='linear'), pnum=pnum_(), title='resize no-aa linear') >>> kwplot.imshow(kwimage.imresize(img, dsize=dsize, antialias=False, interpolation='nearest'), pnum=pnum_(), title='resize no-aa nearest') >>> kwplot.imshow(kwimage.imresize(img, dsize=dsize, antialias=False, interpolation='cubic'), pnum=pnum_(), title='resize no-aa cubic')
Example
>>> # Test single pixel resize >>> import kwimage >>> import numpy as np >>> assert kwimage.imresize(np.random.rand(1, 1, 3), scale=3).shape == (3, 3, 3) >>> assert kwimage.imresize(np.random.rand(1, 1), scale=3).shape == (3, 3)
# cv2.resize(np.random.rand(1, 1, 3), (3, 3))
Todo
[X] When interpolation is area and the number of channels > 4 cv2.resize will error but it is fine for linear interpolation
[ ] TODO: add padding options when letterbox=True
[ ] Allow for pre-clipping when letterbox=True
- kwimage.im_cv2.imscale(img, scale, interpolation=None, return_scale=False)[source]¶
DEPRECATED and removed: use imresize instead
- kwimage.im_cv2.morphology(data, mode, kernel=5, element='rect', iterations=1, border_mode='constant', border_value=0)[source]¶
Executes a morphological operation.
- Parameters:
input (ndarray[dtype=uint8 | float64]) – data (note if mode is hitmiss data must be uint8)
mode (str) – morphology mode, can be one of: ‘erode’, ‘dilate’, ‘open’, ‘close’, ‘gradient’, ‘tophat’, ‘blackhat’, or ‘hitmiss’.
kernel (ndarray | int | Tuple[int, int]) – size of the morphology kernel (w, h) to be constructed according to “element”. If the kernel size is 0, this function returns a copy of the data. Can also be a 2D array which is a custom structuring element. In this case “element” is ignored.
element (str) – structural element, can be ‘rect’, ‘cross’, or ‘ellipse’.
iterations (int) – numer of times to repeat the operation
border_mode (str | int) – Border code or cv2 integer. Border codes are constant (default) replicate, reflect, wrap, reflect101, and transparent.
border_value (int | float | Iterable[int | float]) – Used as the fill value if border_mode is constant. Otherwise this is ignored.
Example
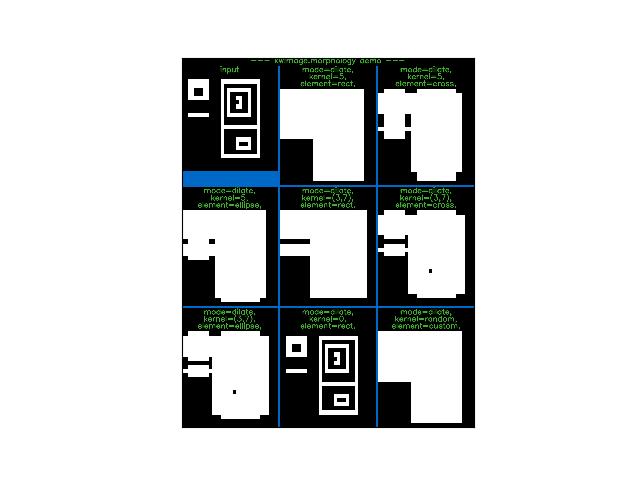
>>> from kwimage.im_cv2 import * # NOQA >>> import kwimage >>> #image = kwimage.grab_test_image(dsize=(380, 380)) >>> image = kwimage.Mask.demo().data * 255 >>> basis = { >>> 'mode': ['dilate'], >>> 'kernel': [5, (3, 7)], >>> 'element': ['rect', 'cross', 'ellipse'], >>> #'mode': ['dilate', 'erode'], >>> } >>> grid = list(ub.named_product(basis)) >>> grid += [{'mode': 'dilate', 'kernel': 0, 'element': 'rect', }] >>> grid += [{'mode': 'dilate', 'kernel': 'random', 'element': 'custom'}] >>> results = {} >>> for params in grid: ... key = ub.urepr(params, compact=1, si=0, nl=1) ... if params['kernel'] == 'random': ... params['kernel'] = np.random.rand(5, 5) ... results[key] = morphology(image, **params) >>> # xdoctest: +REQUIRES(--show) >>> import kwplot >>> kwplot.autompl() >>> to_stack = [] >>> canvas = image >>> canvas = kwimage.imresize(canvas, dsize=(380, 380), interpolation='nearest') >>> canvas = kwimage.draw_header_text(canvas, 'input', color='kitware_green') >>> to_stack.append(canvas) >>> for key, result in results.items(): >>> canvas = result >>> canvas = kwimage.imresize(canvas, dsize=(380, 380), interpolation='nearest') >>> canvas = kwimage.draw_header_text(canvas, key, color='kitware_green') >>> to_stack.append(canvas) >>> canvas = kwimage.stack_images_grid(to_stack, pad=10, bg_value='kitware_blue') >>> canvas = kwimage.draw_header_text(canvas, '--- kwimage.morphology demo ---', color='kitware_green') >>> kwplot.imshow(canvas) >>> kwplot.show_if_requested()
Example
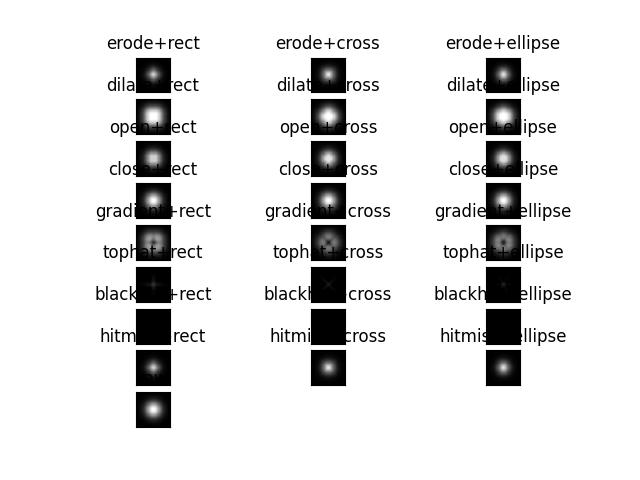
>>> from kwimage.im_cv2 import * # NOQA >>> from kwimage.im_cv2 import _CV2_MORPH_MODES # NOQA >>> from kwimage.im_cv2 import _CV2_STRUCT_ELEMENTS # NOQA >>> #shape = (32, 32) >>> shape = (64, 64) >>> data = (np.random.rand(*shape) > 0.5).astype(np.uint8) >>> import kwimage >>> data = kwimage.gaussian_patch(shape) >>> data = data / data.max() >>> data = kwimage.ensure_uint255(data) >>> results = {} >>> kernel = 5 >>> for mode in _CV2_MORPH_MODES.keys(): ... for element in _CV2_STRUCT_ELEMENTS.keys(): ... results[f'{mode}+{element}'] = morphology(data, mode, kernel=kernel, element=element, iterations=2) >>> results['raw'] = data >>> # xdoctest: +REQUIRES(--show) >>> import kwplot >>> kwplot.autompl() >>> pnum_ = kwplot.PlotNums(nCols=3, nSubplots=len(results)) >>> for k, result in results.items(): >>> kwplot.imshow(result, pnum=pnum_(), title=k) >>> kwplot.show_if_requested()
References
- kwimage.im_cv2.warp_affine(image, transform, dsize=None, antialias=False, interpolation='linear', border_mode=None, border_value=0, large_warp_dim=None, return_info=False)[source]¶
Applies an affine transformation to an image with optional antialiasing.
- Parameters:
image (ndarray) – the input image as a numpy array. Note: this is passed directly to cv2, so it is best to ensure that it is contiguous and using a dtype that cv2 can handle.
transform (ndarray | dict | kwimage.Affine) – a coercable affine matrix. See
kwimage.Affinefor details on what can be coerced.dsize (Tuple[int, int] | None | str) – A integer width and height tuple of the resulting “canvas” image. If None, then the input image size is used.
If specified as a string, dsize is computed based on the given heuristic.
If ‘positive’ (or ‘auto’), dsize is computed such that the positive coordinates of the warped image will fit in the new canvas. In this case, any pixel that maps to a negative coordinate will be clipped. This has the property that the input transformation is not modified. NOTE: there are issues with this when the transformation includes rotation of reflections.
If ‘content’ (or ‘max’), the transform is modified with an extra translation such that both the positive and negative coordinates of the warped image will fit in the new canvas.
antialias (bool) – if True determines if the transform is downsampling and applies antialiasing via gaussian a blur. Defaults to False
interpolation (str | int) – interpolation code or cv2 integer. Interpolation codes are linear, nearest, cubic, lancsoz, and area. Defaults to “linear”.
border_mode (str | int) – Border code or cv2 integer. Border codes are constant (default) replicate, reflect, wrap, reflect101, and transparent.
border_value (int | float | Iterable[int | float]) – Used as the fill value if border_mode is constant. Otherwise this is ignored. Defaults to 0, but can also be defaulted to nan. if border_value is a scalar and there are multiple channels, the value is applied to all channels. More than 4 unique border values for individual channels will cause an error. See OpenCV #22283 for details. In the future we may accept np.ma and return a masked array, but for now that is not implemented.
large_warp_dim (int | None | str) – If specified, perform the warp piecewise in chunks of the specified size. If “auto”, it is set to the maximum “short” value in numpy. This works around a limitation of cv2.warpAffine, which must have image dimensions < SHRT_MAX (=32767 in version 4.5.3)
return_info (bool) – if True, returns information about the operation. In the case where dsize=”content”, this includes the modified transformation.
- Returns:
the warped image, or if return info is True, the warped image and the info dictionary.
- Return type:
ndarray | Tuple[ndarray, Dict]
Todo
- [ ] When dsize=’positive’ but the transform contains an axis flip,
the width / height of the box will become negative. Should we adjust for this?
Example
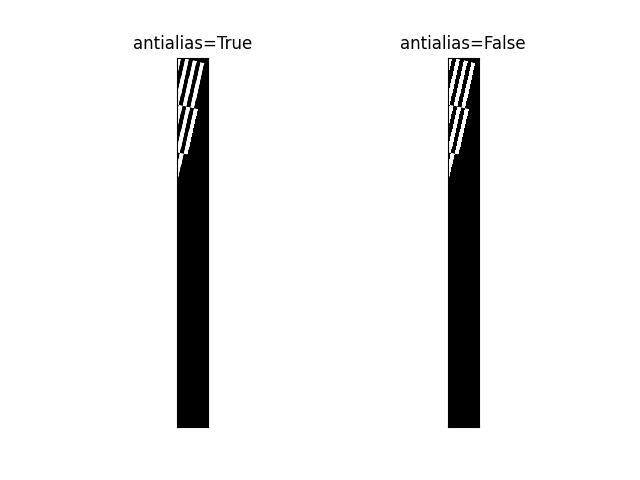
>>> from kwimage.im_cv2 import * # NOQA >>> import kwimage >>> from kwimage.transform import Affine >>> image = kwimage.grab_test_image('astro') >>> #image = kwimage.grab_test_image('checkerboard') >>> transform = Affine.random() @ Affine.scale(0.05) >>> transform = Affine.scale(0.02) >>> warped1 = warp_affine(image, transform, dsize='positive', antialias=1, interpolation='nearest') >>> warped2 = warp_affine(image, transform, dsize='positive', antialias=0) >>> # xdoctest: +REQUIRES(--show) >>> import kwplot >>> kwplot.autompl() >>> pnum_ = kwplot.PlotNums(nRows=1, nCols=2) >>> kwplot.imshow(warped1, pnum=pnum_(), title='antialias=True') >>> kwplot.imshow(warped2, pnum=pnum_(), title='antialias=False') >>> kwplot.show_if_requested()

Example
>>> from kwimage.im_cv2 import * # NOQA >>> import kwimage >>> from kwimage.transform import Affine >>> image = kwimage.grab_test_image('astro') >>> image = kwimage.grab_test_image('checkerboard') >>> transform = Affine.random() @ Affine.scale((.1, 1.2)) >>> warped1 = warp_affine(image, transform, dsize='positive', antialias=1) >>> warped2 = warp_affine(image, transform, dsize='positive', antialias=0) >>> # xdoctest: +REQUIRES(--show) >>> import kwplot >>> kwplot.autompl() >>> pnum_ = kwplot.PlotNums(nRows=1, nCols=2) >>> kwplot.imshow(warped1, pnum=pnum_(), title='antialias=True') >>> kwplot.imshow(warped2, pnum=pnum_(), title='antialias=False') >>> kwplot.show_if_requested()
Example
>>> # Test the case where the input data is empty or the target canvas >>> # is empty, this should be handled like boundary effects >>> import kwimage >>> image = np.random.rand(1, 1, 3) >>> transform = kwimage.Affine.random() >>> result = kwimage.warp_affine(image, transform, dsize=(0, 0)) >>> assert result.shape == (0, 0, 3) >>> # >>> empty_image = np.random.rand(0, 1, 3) >>> result = kwimage.warp_affine(empty_image, transform, dsize=(10, 10)) >>> assert result.shape == (10, 10, 3) >>> # >>> empty_image = np.random.rand(0, 1, 3) >>> result = kwimage.warp_affine(empty_image, transform, dsize=(10, 0)) >>> assert result.shape == (0, 10, 3)
Example
>>> # Demo difference between positive and content dsize >>> from kwimage.im_cv2 import * # NOQA >>> import kwimage >>> from kwimage.transform import Affine >>> image = kwimage.grab_test_image('astro', dsize=(512, 512)) >>> transform = Affine.coerce(offset=(-100, -50), scale=2, theta=0.1) >>> # When warping other images or geometry along with this image >>> # it is important to account for the modified transform when >>> # setting dsize='content'. If dsize='positive', the transform >>> # will remain unchanged wrt other aligned images / geometries. >>> poly = kwimage.Boxes([[350, 5, 130, 290]], 'xywh').to_polygons()[0] >>> # Apply the warping to the images >>> warped_pos, info_pos = warp_affine(image, transform, dsize='positive', return_info=True) >>> warped_con, info_con = warp_affine(image, transform, dsize='content', return_info=True) >>> assert info_pos['dsize'] == (919, 1072) >>> assert info_con['dsize'] == (1122, 1122) >>> assert info_pos['transform'] == transform >>> # Demo the correct and incorrect way to apply transforms >>> poly_pos = poly.warp(transform) >>> poly_con = poly.warp(info_con['transform']) >>> # xdoctest: +REQUIRES(--show) >>> import kwplot >>> kwplot.autompl() >>> # show original >>> kwplot.imshow(image, pnum=(1, 3, 1), title='original') >>> poly.draw(color='green', alpha=0.5, border=True) >>> # show positive warped >>> kwplot.imshow(warped_pos, pnum=(1, 3, 2), title='dsize=positive') >>> poly_pos.draw(color='purple', alpha=0.5, border=True) >>> # show content warped >>> ax = kwplot.imshow(warped_con, pnum=(1, 3, 3), title='dsize=content')[1] >>> poly_con.draw(color='dodgerblue', alpha=0.5, border=True) # correct >>> poly_pos.draw(color='orangered', alpha=0.5, border=True) # incorrect >>> cc = poly_con.to_shapely().centroid >>> cp = poly_pos.to_shapely().centroid >>> ax.text(cc.x, cc.y + 250, 'correctly transformed', color='dodgerblue', >>> backgroundcolor=(0, 0, 0, 0.7), horizontalalignment='center') >>> ax.text(cp.x, cp.y - 250, 'incorrectly transformed', color='orangered', >>> backgroundcolor=(0, 0, 0, 0.7), horizontalalignment='center') >>> kwplot.show_if_requested()

Example
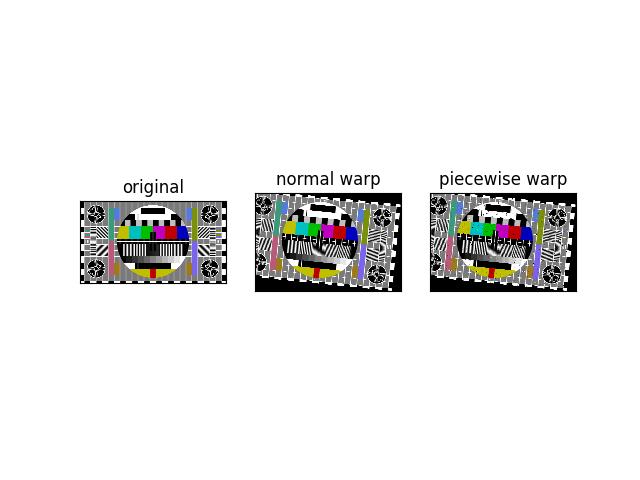
>>> # Demo piecewise transform >>> from kwimage.im_cv2 import * # NOQA >>> import kwimage >>> from kwimage.transform import Affine >>> image = kwimage.grab_test_image('pm5644') >>> transform = Affine.coerce(offset=(-100, -50), scale=2, theta=0.1) >>> warped_piecewise, info = warp_affine(image, transform, dsize='positive', return_info=True, large_warp_dim=32) >>> warped_normal, info = warp_affine(image, transform, dsize='positive', return_info=True, large_warp_dim=None) >>> # xdoctest: +REQUIRES(--show) >>> import kwplot >>> kwplot.autompl() >>> kwplot.imshow(image, pnum=(1, 3, 1), title='original') >>> kwplot.imshow(warped_normal, pnum=(1, 3, 2), title='normal warp') >>> kwplot.imshow(warped_piecewise, pnum=(1, 3, 3), title='piecewise warp')
Example
>>> from kwimage.im_cv2 import * # NOQA >>> import kwimage >>> # TODO: Explain why the bottom left is interpolated with 0's >>> # And not 2s, probably has to do with interpretation of pixels >>> # as points and not areas. >>> image = np.full((6, 6), fill_value=3, dtype=np.uint8) >>> transform = kwimage.Affine.eye() >>> transform = kwimage.Affine.coerce(offset=.5) @ transform >>> transform = kwimage.Affine.coerce(scale=2) @ transform >>> warped = kwimage.warp_affine(image, transform, dsize=(12, 12))
Example
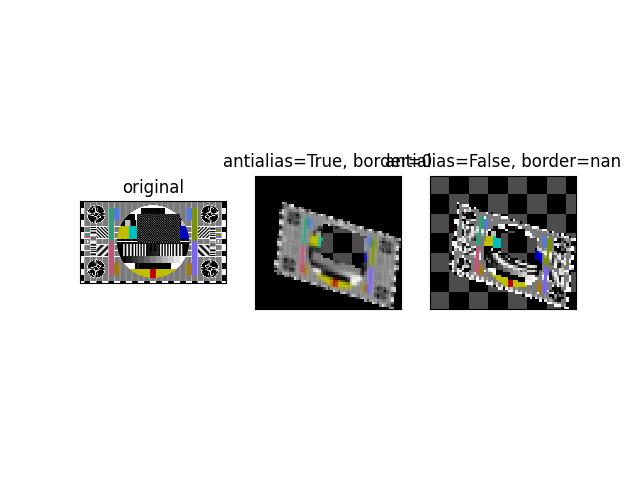
>>> # Demo how nans are handled >>> from kwimage.im_cv2 import * # NOQA >>> import kwimage >>> image = kwimage.grab_test_image('pm5644') >>> image = kwimage.ensure_float01(image) >>> image[100:300, 400:700] = np.nan >>> transform = kwimage.Affine.coerce(scale=0.05, offset=10.5, theta=0.3, shearx=0.2) >>> warped1 = warp_affine(image, transform, dsize='positive', antialias=1, interpolation='linear', border_value=0) >>> warped2 = warp_affine(image, transform, dsize='positive', antialias=0, border_value=np.nan) >>> assert np.isnan(warped1).any() >>> assert np.isnan(warped2).any() >>> assert warped1[np.isnan(warped1).any(axis=2)].all() >>> assert warped2[np.isnan(warped2).any(axis=2)].all() >>> print('warped1.shape = {!r}'.format(warped1.shape)) >>> print('warped2.shape = {!r}'.format(warped2.shape)) >>> assert warped2.shape == warped1.shape >>> warped2[np.isnan(warped2).any(axis=2)] >>> # xdoctest: +REQUIRES(--show) >>> import kwplot >>> kwplot.autompl() >>> pnum_ = kwplot.PlotNums(nRows=1, nCols=3) >>> image_canvas = kwimage.fill_nans_with_checkers(image) >>> warped1_canvas = kwimage.fill_nans_with_checkers(warped1) >>> warped2_canvas = kwimage.fill_nans_with_checkers(warped2) >>> kwplot.imshow(image_canvas, pnum=pnum_(), title='original') >>> kwplot.imshow(warped1_canvas, pnum=pnum_(), title='antialias=True, border=0') >>> kwplot.imshow(warped2_canvas, pnum=pnum_(), title='antialias=False, border=nan') >>> kwplot.show_if_requested()
Example
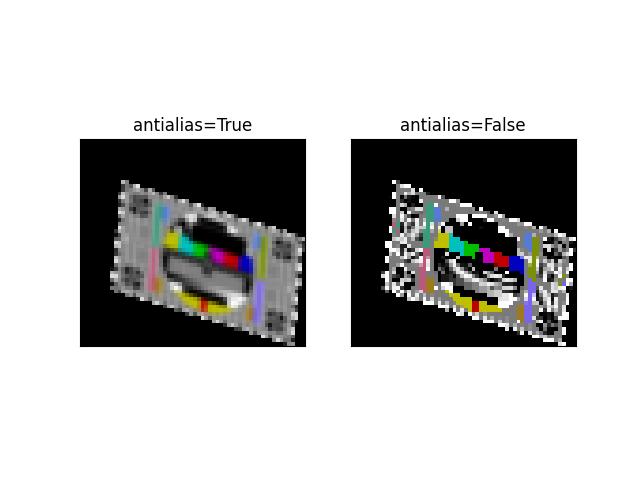
>>> # Demo how of how we also handle masked arrays >>> from kwimage.im_cv2 import * # NOQA >>> import kwimage >>> _image = kwimage.grab_test_image('pm5644') >>> _image = kwimage.ensure_float01(_image) >>> _image[100:200, 400:700] = np.nan >>> mask = np.isnan(_image) >>> data = np.nan_to_num(_image) >>> image = np.ma.MaskedArray(data=data, mask=mask) >>> transform = kwimage.Affine.coerce(scale=0.05, offset=10.5, theta=0.3, shearx=0.2) >>> warped1 = warp_affine(image, transform, dsize='positive', antialias=1, interpolation='linear') >>> assert isinstance(warped1, np.ma.MaskedArray) >>> warped2 = warp_affine(image, transform, dsize='positive', antialias=0) >>> print('warped1.shape = {!r}'.format(warped1.shape)) >>> print('warped2.shape = {!r}'.format(warped2.shape)) >>> assert warped2.shape == warped1.shape >>> # xdoctest: +REQUIRES(--show) >>> import kwplot >>> kwplot.autompl() >>> pnum_ = kwplot.PlotNums(nRows=1, nCols=2) >>> kwplot.imshow(warped1, pnum=pnum_(), title='antialias=True') >>> kwplot.imshow(warped2, pnum=pnum_(), title='antialias=False') >>> kwplot.show_if_requested()
- kwimage.im_cv2.warp_image(image, transform, dsize=None, antialias=False, interpolation='linear', border_mode=None, border_value=0, large_warp_dim=None, return_info=False)[source]¶
Applies an transformation to an image with optional antialiasing.
- Parameters:
image (ndarray) – the input image as a numpy array. Note: this is passed directly to cv2, so it is best to ensure that it is contiguous and using a dtype that cv2 can handle.
transform (ndarray | dict | kwimage.Matrix) – a coercable affine or projective matrix. See
kwimage.Affineandkwimage.Projectivefor details on what can be coerced.dsize (Tuple[int, int] | None | str) – A integer width and height tuple of the resulting “canvas” image. If None, then the input image size is used.
If specified as a string, dsize is computed based on the given heuristic.
If ‘positive’ (or ‘auto’), dsize is computed such that the positive coordinates of the warped image will fit in the new canvas. In this case, any pixel that maps to a negative coordinate will be clipped. This has the property that the input transformation is not modified.
If ‘content’ (or ‘max’), the transform is modified with an extra translation such that both the positive and negative coordinates of the warped image will fit in the new canvas.
antialias (bool) – if True determines if the transform is downsampling and applies antialiasing via gaussian a blur. Defaults to False
interpolation (str | int) – interpolation code or cv2 integer. Interpolation codes are linear, nearest, cubic, lancsoz, and area. Defaults to “linear”.
border_mode (str | int) – Border code or cv2 integer. Border codes are constant (default) replicate, reflect, wrap, reflect101, and transparent.
border_value (int | float | Iterable[int | float]) – Used as the fill value if border_mode is constant. Otherwise this is ignored. Defaults to 0, but can also be defaulted to nan. if border_value is a scalar and there are multiple channels, the value is applied to all channels. More than 4 unique border values for individual channels will cause an error. See OpenCV #22283 for details. In the future we may accept np.ma and return a masked array, but for now that is not implemented.
large_warp_dim (int | None | str) – If specified, perform the warp piecewise in chunks of the specified size. If “auto”, it is set to the maximum “short” value in numpy. This works around a limitation of cv2.warpAffine, which must have image dimensions < SHRT_MAX (=32767 in version 4.5.3)
return_info (bool) – if True, returns information about the operation. In the case where dsize=”content”, this includes the modified transformation.
- Returns:
the warped image, or if return info is True, the warped image and the info dictionary.
- Return type:
ndarray | Tuple[ndarray, Dict]
Example
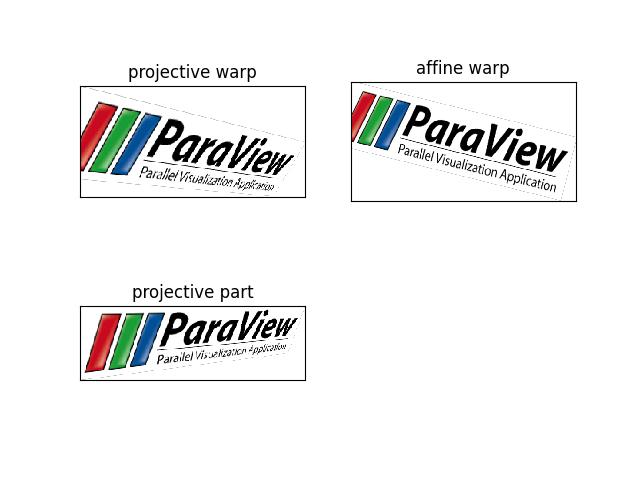
>>> from kwimage.im_cv2 import * # NOQA >>> import kwimage >>> image = kwimage.grab_test_image('paraview') >>> tf_homog = kwimage.Projective.random(rng=30342110) @ kwimage.Projective.coerce(uv=[0.001, 0.001]) >>> tf_aff = kwimage.Affine.coerce(ub.udict(tf_homog.decompose()) - {'uv'}) >>> tf_uv = kwimage.Projective.coerce(ub.udict(tf_homog.decompose()) & {'uv'}) >>> warped1 = kwimage.warp_image(image, tf_homog, dsize='positive') >>> warped2 = kwimage.warp_image(image, tf_aff, dsize='positive') >>> warped3 = kwimage.warp_image(image, tf_uv, dsize='positive') >>> # xdoctest: +REQUIRES(--show) >>> import kwplot >>> kwplot.autompl() >>> pnum_ = kwplot.PlotNums(nRows=2, nCols=2) >>> kwplot.imshow(warped1, pnum=pnum_(), title='projective warp') >>> kwplot.imshow(warped2, pnum=pnum_(), title='affine warp') >>> kwplot.imshow(warped3, pnum=pnum_(), title='projective part') >>> kwplot.show_if_requested()
- kwimage.im_cv2.warp_projective(image, transform, dsize=None, antialias=False, interpolation='linear', border_mode=None, border_value=0, large_warp_dim=None, return_info=False)[source]¶
Applies an projective transformation to an image with optional antialiasing.
- Parameters:
image (ndarray) – the input image as a numpy array. Note: this is passed directly to cv2, so it is best to ensure that it is contiguous and using a dtype that cv2 can handle.
transform (ndarray | dict | kwimage.Projective) – a coercable projective matrix. See
kwimage.Projectivefor details on what can be coerced.dsize (Tuple[int, int] | None | str) – A integer width and height tuple of the resulting “canvas” image. If None, then the input image size is used.
If specified as a string, dsize is computed based on the given heuristic.
If ‘positive’ (or ‘auto’), dsize is computed such that the positive coordinates of the warped image will fit in the new canvas. In this case, any pixel that maps to a negative coordinate will be clipped. This has the property that the input transformation is not modified.
If ‘content’ (or ‘max’), the transform is modified with an extra translation such that both the positive and negative coordinates of the warped image will fit in the new canvas.
antialias (bool) – if True determines if the transform is downsampling and applies antialiasing via gaussian a blur. Defaults to False
interpolation (str | int) – interpolation code or cv2 integer. Interpolation codes are linear, nearest, cubic, lancsoz, and area. Defaults to “linear”.
border_mode (str | int) – Border code or cv2 integer. Border codes are constant (default) replicate, reflect, wrap, reflect101, and transparent.
border_value (int | float | Iterable[int | float]) – Used as the fill value if border_mode is constant. Otherwise this is ignored. Defaults to 0, but can also be defaulted to nan. if border_value is a scalar and there are multiple channels, the value is applied to all channels. More than 4 unique border values for individual channels will cause an error. See OpenCV #22283 for details. In the future we may accept np.ma and return a masked array, but for now that is not implemented.
large_warp_dim (int | None | str) – If specified, perform the warp piecewise in chunks of the specified size. If “auto”, it is set to the maximum “short” value in numpy. This works around a limitation of cv2.warpAffine, which must have image dimensions < SHRT_MAX (=32767 in version 4.5.3)
return_info (bool) – if True, returns information about the operation. In the case where dsize=”content”, this includes the modified transformation.
- Returns:
the warped image, or if return info is True, the warped image and the info dictionary.
- Return type:
ndarray | Tuple[ndarray, Dict]