kwimage.structs.detections module¶
Structure for efficient access and modification of bounding boxes with associated scores and class labels. Builds on top of the kwimage.Boxes structure.
Also can optionally incorporate kwimage.PolygonList for segmentation masks and kwimage.PointsList for keypoints.
- If you want to visualize boxes and scores you can do this:
>>> # Given data >>> data = np.random.rand(10, 4) * 224 >>> scores = np.random.rand(10,) >>> class_idxs = np.random.randint(0, 3, size=10) >>> classes = ['class1', 'class2', 'class3'] >>> # >>> # Wrap your data with a Detections object >>> import kwimage >>> dets = kwimage.Detections( >>> boxes=kwimage.Boxes(data, format='xywh'), >>> scores=scores, >>> class_idxs=class_idxs, >>> classes=classes, >>> ) >>> # xdoctest: +REQUIRES(module:kwplot) >>> dets.draw() >>> import matplotlib.pyplot as plt >>> plt.gca().set_xlim(0, 224) >>> plt.gca().set_ylim(0, 224)
- class kwimage.structs.detections._DetDrawMixin[source]¶
Bases:
objectNon critical methods for visualizing detections
- draw(color='blue', alpha=None, labels=True, centers=False, lw=2, fill=False, ax=None, radius=5, kpts=True, sseg=True, setlim=False, boxes=True)[source]¶
Draws boxes using matplotlib
Example
>>> # xdoctest: +REQUIRES(module:kwplot) >>> self = Detections.random(num=10, scale=512.0, rng=0, classes=['a', 'b', 'c']) >>> self.boxes.translate((-128, -128), inplace=True) >>> image = (np.random.rand(256, 256) * 255).astype(np.uint8) >>> # xdoctest: +REQUIRES(--show) >>> import kwplot >>> kwplot.autompl() >>> fig = kwplot.figure(fnum=1, doclf=True) >>> kwplot.imshow(image) >>> # xdoctest: +REQUIRES(--show) >>> self.draw(color='blue', alpha=None) >>> # xdoctest: +REQUIRES(--show) >>> for o in fig.findobj(): # http://matplotlib.1069221.n5.nabble.com/How-to-turn-off-all-clipping-td1813.html >>> o.set_clip_on(False) >>> kwplot.show_if_requested()
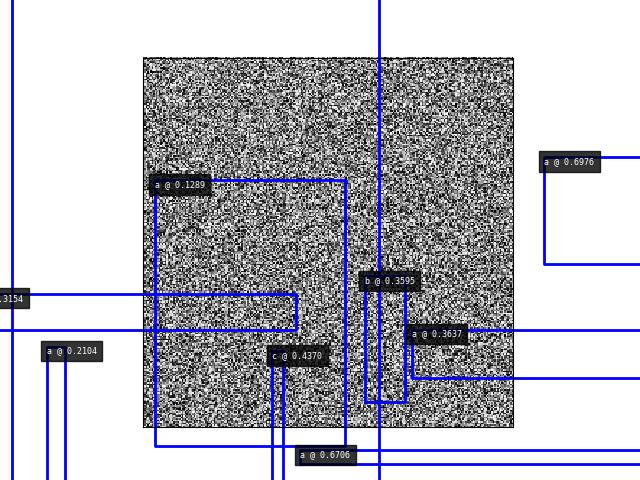
- draw_on(image=None, color='blue', alpha=None, labels=True, radius=5, kpts=True, sseg=True, boxes=True, ssegkw=None, label_loc='top_left', thickness=2)[source]¶
Draws boxes directly on the image using OpenCV
- Parameters:
image (ndarray[Any, UInt8]) – must be in uint8 format
color (str | Any | List[Any]) – one color for all boxes or a list of colors for each box. Or the string “classes”, in which case it will use a different color for each class (specified in the classes object if possible). Extended types: str | ColorLike | List[ColorLike]
alpha (float) – Transparency of overlay. can be a scalar or a list for each box
labels (bool | str | List[str]) – if True, use categorie names as the labels. See _make_labels for details. Otherwise a manually specified text label for each box.
boxes (bool) – if True draw the boxes
kpts (bool) – if True draw the keypoints
sseg (bool) – if True draw the segmentations
ssegkw (dict) – extra arguments passed to segmentations.draw_on
radius (float) – passed to keypoints.draw_on
label_loc (str) – indicates where labels (if specified) should be drawn. passed to boxes.draw_on
thickness (int) – rectangle thickness, negative values will draw a filled rectangle. passed to boxes.draw_on. Defaults to 2.
- Returns:
image with labeled boxes drawn on it
- Return type:
ndarray[Any, UInt8]
CommandLine
xdoctest -m kwimage.structs.detections _DetDrawMixin.draw_on:1 --profile --show
Example
>>> # xdoctest: +REQUIRES(module:kwplot) >>> import kwimage >>> import kwplot >>> self = kwimage.Detections.random(num=10, scale=512, rng=0) >>> image = (np.random.rand(512, 512) * 255).astype(np.uint8) >>> image2 = self.draw_on(image, color='blue') >>> # xdoctest: +REQUIRES(--show) >>> kwplot.autompl() >>> kwplot.figure(fnum=2000, doclf=True) >>> kwplot.imshow(image2) >>> kwplot.show_if_requested()

Example
>>> # xdoctest: +REQUIRES(module:kwplot) >>> import kwimage >>> from kwimage.structs.detections import * # NOQA >>> import kwplot >>> self = kwimage.Detections.random(num=10, scale=512, rng=0) >>> image = (np.random.rand(512, 512) * 255).astype(np.uint8) >>> image2 = self.draw_on(image, color='classes') >>> # xdoctest: +REQUIRES(--show) >>> kwplot.autompl() >>> kwplot.figure(fnum=2000, doclf=True) >>> kwplot.imshow(image2) >>> kwplot.show_if_requested()

Example
>>> # xdoctest: +REQUIRES(module:kwplot) >>> # xdoctest: +REQUIRES(--profile) >>> import kwimage >>> import kwplot >>> self = kwimage.Detections.random(num=100, scale=512, rng=0, keypoints=True, segmentations=True) >>> image = (np.random.rand(512, 512) * 255).astype(np.uint8) >>> image2 = self.draw_on(image, color='blue') >>> # xdoctest: +REQUIRES(--show) >>> kwplot.figure(fnum=2000, doclf=True) >>> kwplot.autompl() >>> kwplot.imshow(image2) >>> kwplot.show_if_requested()
Example
>>> # xdoctest: +REQUIRES(module:kwplot) >>> # Test that boxes with tiny scores are drawn correctly >>> import kwimage >>> import kwplot >>> self = kwimage.Detections.random(num=10, scale=512, rng=0) >>> self.data['scores'][:] = 1.23e-8 >>> image = (np.random.rand(512, 512) * 255).astype(np.uint8) >>> image2 = self.draw_on(image, color='blue') >>> # xdoctest: +REQUIRES(--show) >>> kwplot.autompl() >>> kwplot.figure(fnum=2000, doclf=True) >>> kwplot.imshow(image2) >>> kwplot.show_if_requested()
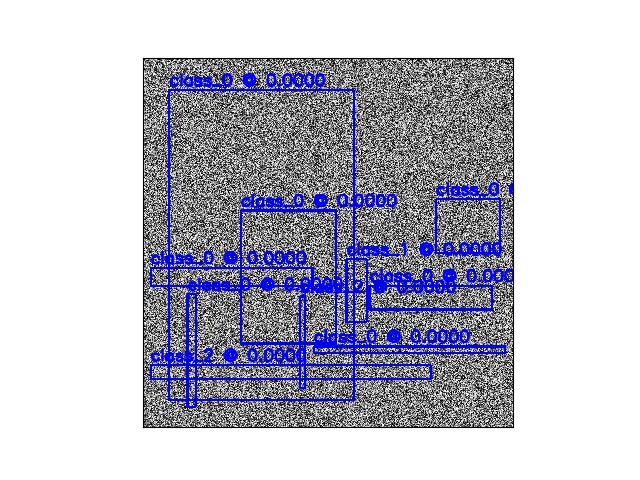
- _make_colors(color)[source]¶
Handles special settings of color.
If color == ‘classes’, then choose a distinct color for each category
- class kwimage.structs.detections._DetAlgoMixin[source]¶
Bases:
objectNon critical methods for algorithmic manipulation of detections
- non_max_supression(thresh=0.0, perclass=False, impl='auto', daq=False, device_id=None)[source]¶
Find high scoring minimally overlapping detections
- Parameters:
thresh (float) – iou threshold between 0 and 1. A box is removed if it overlaps with a previously chosen box by more than this threshold. Higher values are are more permissive (more boxes are returned). A value of 0 means that returned boxes will have no overlap.
perclass (bool) – if True, works on a per-class basis
impl (str) – nms implementation to use
daq (bool | Dict) – if False, uses reqgular nms, otherwise uses divide and conquor algorithm. If daq is a Dict, then it is used as the kwargs to kwimage.daq_spatial_nms
device_id – try not to use. only used if impl is gpu
- Returns:
indices of boxes to keep
- Return type:
ndarray[Shape[‘*’], Integer]
Example
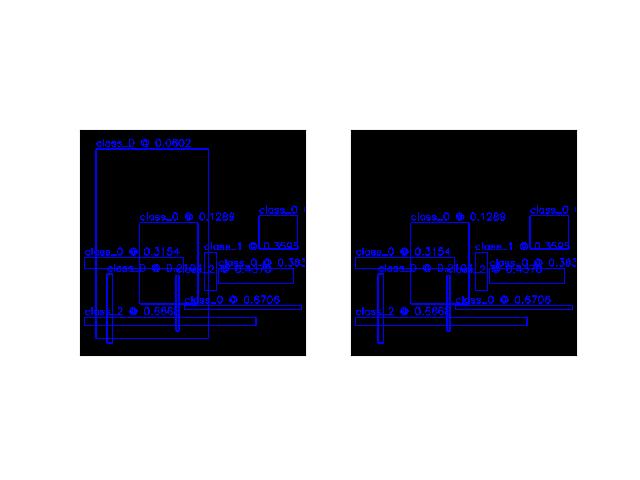
>>> import kwimage >>> dets1 = kwimage.Detections.random(rng=0).scale((512, 512)) >>> keep = dets1.non_max_supression(thresh=0.2) >>> dets2 = dets1.take(keep) >>> # xdoctest: +REQUIRES(--show) >>> import kwplot >>> kwplot.autompl() >>> canvas = np.zeros((512, 512, 3)) >>> canvas1 = dets1.draw_on(canvas.copy()) >>> canvas2 = dets2.draw_on(canvas.copy()) >>> kwplot.figure(fnum=1, pnum=(1, 2, 1)) >>> kwplot.imshow(canvas1) >>> kwplot.figure(fnum=1, pnum=(1, 2, 2)) >>> kwplot.imshow(canvas2)
- non_max_supress(thresh=0.0, perclass=False, impl='auto', daq=False)[source]¶
Convinience method. Like non_max_supression, but returns to supressed boxes instead of the indices to keep.
- rasterize(bg_size, input_dims, soften=1, tf_data_to_img=None, img_dims=None, exclude=[])[source]¶
Ambiguous conversion from a Heatmap to a Detections object.
- SeeAlso:
Heatmap.detect
- Returns:
raster-space detections.
- Return type:
Example
>>> # xdoctest: +REQUIRES(module:ndsampler) >>> from kwimage.structs.detections import * # NOQA >>> self, iminfo, sampler = Detections.demo() >>> image = iminfo['imdata'][:] >>> input_dims = iminfo['imdata'].shape[0:2] >>> bg_size = [100, 100] >>> heatmap = self.rasterize(bg_size, input_dims) >>> # xdoctest: +REQUIRES(--show) >>> import kwplot >>> kwplot.autompl() >>> kwplot.figure(fnum=1, pnum=(2, 2, 1)) >>> heatmap.draw(invert=True) >>> kwplot.figure(fnum=1, pnum=(2, 2, 2)) >>> kwplot.imshow(heatmap.draw_on(image)) >>> kwplot.figure(fnum=1, pnum=(2, 1, 2)) >>> kwplot.imshow(heatmap.draw_stacked())
- class kwimage.structs.detections.Detections(data=None, meta=None, datakeys=None, metakeys=None, checks=True, **kwargs)[source]¶
Bases:
NiceRepr,_DetAlgoMixin,_DetDrawMixinContainer for holding and manipulating multiple detections.
- Variables:
data (Dict) –
dictionary containing corresponding lists. The length of each list is the number of detections. This contains the bounding boxes, confidence scores, and class indices. Details of the most common keys and types are as follows:
boxes (kwimage.Boxes[ArrayLike]): multiple bounding boxes scores (ArrayLike): associated scores class_idxs (ArrayLike): associated class indices segmentations (ArrayLike): segmentations masks for each box, members can be
MaskorMultiPolygon. keypoints (ArrayLike): keypoints for each box. Members should bePoints.Additional custom keys may be specified as long as (a) the values are array-like and the first axis corresponds to the standard data values and (b) are custom keys are listed in the datakeys kwargs when constructing the Detections.
meta (Dict) – This contains contextual information about the detections. This includes the class names, which can be indexed into via the class indexes.
Example
>>> import kwimage >>> dets = kwimage.Detections( >>> # there are expected keys that do not need registration >>> boxes=kwimage.Boxes.random(3), >>> class_idxs=[0, 1, 1], >>> classes=['a', 'b'], >>> # custom data attrs must align with boxes >>> myattr1=np.random.rand(3), >>> myattr2=np.random.rand(3, 2, 8), >>> # there are no restrictions on metadata >>> mymeta='a custom metadata string', >>> # Note that any key not in kwimage.Detections.__datakeys__ or >>> # kwimage.Detections.__metakeys__ must be registered at the >>> # time of construction. >>> datakeys=['myattr1', 'myattr2'], >>> metakeys=['mymeta'], >>> checks=True, >>> ) >>> print('dets = {}'.format(dets)) dets = <Detections(3)>
Construct a Detections object by either explicitly specifying the internal data and meta dictionary structures or by passing expected attribute names as kwargs.
- Parameters:
data (Dict[str, ArrayLike]) – explicitly specify the data dictionary
meta (Dict[str, object]) – explicitly specify the meta dictionary
datakeys (List[str]) – a list of custom attributes that should be considered as data (i.e. must be an array aligned with boxes).
metakeys (List[str]) – a list of custom attributes that should be considered as metadata (i.e. can be arbitrary).
checks (bool) – if True and arguments are passed by kwargs, then check / ensure that all types are compatible. Defaults to True.
**kwargs – specify any key for the data or meta dictionaries.
Note
Custom data and metadata can be specified as long as you pass the names of these keys in the datakeys and/or metakeys kwargs.
In the case where you specify a custom attribute as a list, it will “currently” (we may change this behavior in the future) be coerced into a numpy or torch array. If you want to store a generic Python list, wrap the custom list in a
_generic.ObjectList.Example
>>> # Coerce to numpy >>> import kwimage >>> dets = Detections( >>> boxes=kwimage.Boxes.random(3).numpy(), >>> class_idxs=[0, 1, 1], >>> checks=True, >>> ) >>> # xdoctest: +REQUIRES(module:torch) >>> # Coerce to tensor >>> dets = Detections( >>> boxes=kwimage.Boxes.random(3).tensor(), >>> class_idxs=[0, 1, 1], >>> checks=True, >>> ) >>> # Error on incompatible types >>> import pytest >>> with pytest.raises(TypeError): >>> dets = Detections( >>> boxes=kwimage.Boxes.random(3).tensor(), >>> scores=np.random.rand(3), >>> class_idxs=[0, 1, 1], >>> checks=True, >>> )
Example
>>> self = Detections.random(10) >>> other = Detections(self) >>> assert other.data == self.data >>> assert other.data is self.data, 'try not to copy unless necessary'
- classmethod coerce(data=None, **kwargs)[source]¶
The “try-anything to get what I want” constructor
- Parameters:
data
**kwargs – currently boxes and cnames
Example
>>> from kwimage.structs.detections import * # NOQA >>> import kwimage >>> kwargs = dict( >>> boxes=kwimage.Boxes.random(4), >>> cnames=['a', 'b', 'c', 'c'], >>> ) >>> data = {} >>> self = kwimage.Detections.coerce(data, **kwargs)
- classmethod from_coco_annots(anns, cats=None, classes=None, kp_classes=None, shape=None, dset=None)[source]¶
Create a Detections object from a list of coco-like annotations.
- Parameters:
anns (List[Dict]) – list of coco-like annotation objects
dset (kwcoco.CocoDataset) – if specified, cats, classes, and kp_classes can are ignored.
cats (List[Dict]) – coco-format category information. Used only if dset is not specified.
classes (kwcoco.CategoryTree) – category tree with coco class info. Used only if dset is not specified.
kp_classes (kwcoco.CategoryTree) – keypoint category tree with coco keypoint class info. Used only if dset is not specified.
shape (tuple) – shape of parent image
- Returns:
a detections object
- Return type:
Example
>>> from kwimage.structs.detections import * # NOQA >>> # xdoctest: +REQUIRES(--module:ndsampler) >>> anns = [{ >>> 'id': 0, >>> 'image_id': 1, >>> 'category_id': 2, >>> 'bbox': [2, 3, 10, 10], >>> 'keypoints': [4.5, 4.5, 2], >>> 'segmentation': { >>> 'counts': '_11a04M2O0O20N101N3L_5', >>> 'size': [20, 20], >>> }, >>> }] >>> dataset = { >>> 'images': [], >>> 'annotations': [], >>> 'categories': [ >>> {'id': 0, 'name': 'background'}, >>> {'id': 2, 'name': 'class1', 'keypoints': ['spot']} >>> ] >>> } >>> #import ndsampler >>> #dset = ndsampler.CocoDataset(dataset) >>> cats = dataset['categories'] >>> dets = Detections.from_coco_annots(anns, cats)
Example
>>> # xdoctest: +REQUIRES(--module:ndsampler) >>> # Test case with no category information >>> from kwimage.structs.detections import * # NOQA >>> anns = [{ >>> 'id': 0, >>> 'image_id': 1, >>> 'category_id': None, >>> 'bbox': [2, 3, 10, 10], >>> 'prob': [.1, .9], >>> }] >>> cats = [ >>> {'id': 0, 'name': 'background'}, >>> {'id': 2, 'name': 'class1'} >>> ] >>> dets = Detections.from_coco_annots(anns, cats)
Example
>>> import kwimage >>> # xdoctest: +REQUIRES(--module:ndsampler) >>> import ndsampler >>> sampler = ndsampler.CocoSampler.demo('photos') >>> iminfo, anns = sampler.load_image_with_annots(1) >>> shape = iminfo['imdata'].shape[0:2] >>> kp_classes = sampler.dset.keypoint_categories() >>> dets = kwimage.Detections.from_coco_annots( >>> anns, sampler.dset.dataset['categories'], sampler.catgraph, >>> kp_classes, shape=shape)
- to_coco(cname_to_cat=None, style='orig', image_id=None, dset=None)[source]¶
Converts this set of detections into coco-like annotation dictionaries.
Note
Not all aspects of the MS-COCO format can be accurately represented, so some liberties are taken. The MS-COCO standard defines that annotations should specifiy a category_id field, but in some cases this information is not available so we will populate a ‘category_name’ field if possible and in the worst case fall back to ‘category_index’.
Additionally, detections may contain additional information beyond the MS-COCO standard, and this information (e.g. weight, prob, score) is added as forign fields.
- Parameters:
cname_to_cat – currently ignored.
style (str) – either ‘orig’ (for the original coco format) or ‘new’ for the more general kwcoco-style coco format. Defaults to ‘orig’
image_id (int) – if specified, populates the image_id field of each image.
dset (kwcoco.CocoDataset | None) – if specified, attempts to populate the category_id field to be compatible with this coco dataset.
- Yields:
dict – coco-like annotation structures
Example
>>> # xdoctest: +REQUIRES(module:ndsampler) >>> from kwimage.structs.detections import * >>> self = Detections.demo()[0] >>> cname_to_cat = None >>> list(self.to_coco())
- property boxes¶
- property class_idxs¶
- property scores¶
typically only populated for predicted detections
- property probs¶
typically only populated for predicted detections
- property weights¶
typically only populated for groundtruth detections
- property classes¶
- warp(transform, input_dims=None, output_dims=None, inplace=False)[source]¶
Spatially warp the detections.
- Parameters:
transform (kwimage.Affine | ndarray | Callable | Any) – Something coercable to a transform. Usually a kwimage.Affine object
input_dims (Tuple[int, int]) – shape of the expected input canvas
output_dims (Tuple[int, int]) – shape of the expected output canvas
inplace (bool) – if true operate inplace
- Returns:
the warped detections object
- Return type:
Example
>>> import skimage >>> transform = skimage.transform.AffineTransform(scale=(2, 3), translation=(4, 5)) >>> self = Detections.random(2) >>> new = self.warp(transform) >>> assert new.boxes == self.boxes.warp(transform) >>> assert new != self
- scale(factor, output_dims=None, inplace=False)[source]¶
Spatially scale the detections.
Example
>>> import skimage >>> transform = skimage.transform.AffineTransform(scale=(2, 3), translation=(4, 5)) >>> self = Detections.random(2) >>> new = self.warp(transform) >>> assert new.boxes == self.boxes.warp(transform) >>> assert new != self
- translate(offset, output_dims=None, inplace=False)[source]¶
Spatially translate the detections.
Example
>>> import skimage >>> self = Detections.random(2) >>> new = self.translate(10)
- classmethod concatenate(dets)[source]¶
- Parameters:
boxes (Sequence[Detections]) – list of detections to concatenate
- Returns:
stacked detections
- Return type:
Example
>>> self = Detections.random(2) >>> other = Detections.random(3) >>> dets = [self, other] >>> new = Detections.concatenate(dets) >>> assert new.num_boxes() == 5
>>> self = Detections.random(2, segmentations=True) >>> other = Detections.random(3, segmentations=True) >>> dets = [self, other] >>> new = Detections.concatenate(dets) >>> assert new.num_boxes() == 5
- argsort(reverse=True)[source]¶
Sorts detection indices by descending (or ascending) scores
- Returns:
sorted indices torch.Tensor: sorted indices if using torch backends
- Return type:
ndarray[Shape[‘*’], Integer]
- sort(reverse=True)[source]¶
Sorts detections by descending (or ascending) scores
- Returns:
sorted copy of self
- Return type:
- compress(flags, axis=0)[source]¶
Returns a subset where corresponding locations are True.
- Parameters:
flags (ndarray[Any, Bool] | torch.Tensor) – mask marking selected items
- Returns:
subset of self
- Return type:
CommandLine
xdoctest -m kwimage.structs.detections Detections.compress
Example
>>> # xdoctest: +REQUIRES(module:torch) >>> import kwimage >>> dets = kwimage.Detections.random(keypoints='dense') >>> flags = np.random.rand(len(dets)) > 0.5 >>> subset = dets.compress(flags) >>> assert len(subset) == flags.sum() >>> subset = dets.tensor().compress(flags) >>> assert len(subset) == flags.sum()
- take(indices, axis=0)[source]¶
Returns a subset specified by indices
- Parameters:
indices (ndarray[Any, Integer]) – indices to select
- Returns:
subset of self
- Return type:
Example
>>> import kwimage >>> dets = kwimage.Detections(boxes=kwimage.Boxes.random(10)) >>> subset = dets.take([2, 3, 5, 7]) >>> assert len(subset) == 4 >>> # xdoctest: +REQUIRES(module:torch) >>> subset = dets.tensor().take([2, 3, 5, 7]) >>> assert len(subset) == 4
- property device¶
If the backend is torch returns the data device, otherwise None
- numpy()[source]¶
Converts tensors to numpy. Does not change memory if possible.
Example
>>> # xdoctest: +REQUIRES(module:torch) >>> self = Detections.random(3).tensor() >>> newself = self.numpy() >>> self.scores[0] = 0 >>> assert newself.scores[0] == 0 >>> self.scores[0] = 1 >>> assert self.scores[0] == 1 >>> self.numpy().numpy()
- property dtype¶
- tensor(device=NoParam)[source]¶
Converts numpy to tensors. Does not change memory if possible.
Example
>>> # xdoctest: +REQUIRES(module:torch) >>> from kwimage.structs.detections import * >>> self = Detections.random(3) >>> newself = self.tensor() >>> self.scores[0] = 0 >>> assert newself.scores[0] == 0 >>> self.scores[0] = 1 >>> assert self.scores[0] == 1 >>> self.tensor().tensor()
- classmethod random(num=10, scale=1.0, classes=3, keypoints=False, segmentations=False, tensor=False, rng=None)[source]¶
Creates dummy data, suitable for use in tests and benchmarks
- Parameters:
num (int) – number of boxes
scale (float | tuple) – bounding image size. Defaults to 1.0
classes (int | Sequence) – list of class labels or number of classes
keypoints (bool) – if True include random keypoints for each box. Defaults to False.
segmentations (bool) – if True include random segmentations for each box. Defaults to False.
tensor (bool) – determines backend. DEPRECATED. Call
.tensor()on resulting object instead.rng (int | RandomState | None) – random state or seed
- Returns:
random detections
- Return type:
Example
>>> import kwimage >>> dets = kwimage.Detections.random(keypoints='jagged') >>> dets.data['keypoints'].data[0].data >>> dets.data['keypoints'].meta >>> dets = kwimage.Detections.random(keypoints='dense') >>> dets = kwimage.Detections.random(keypoints='dense', segmentations=True).scale(1000) >>> # xdoctest:+REQUIRES(--show) >>> import kwplot >>> kwplot.autompl() >>> dets.draw(setlim=True)
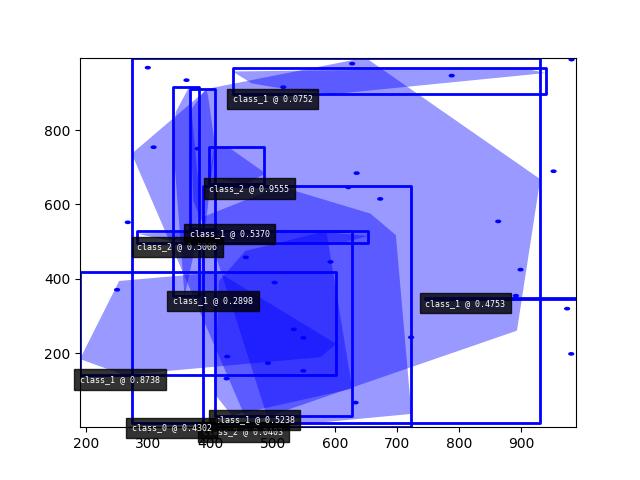
Example
>>> import kwimage >>> dets = kwimage.Detections.random( >>> keypoints='jagged', segmentations=True, rng=0).scale(1000) >>> print('dets = {}'.format(dets)) dets = <Detections(10)> >>> dets.data['boxes'].quantize(inplace=True) >>> print('dets.data = {}'.format(ub.urepr( >>> dets.data, nl=1, with_dtype=False, strvals=True, sort=1))) dets.data = { 'boxes': <Boxes(xywh, array([[548, 544, 55, 172], [423, 645, 15, 247], [791, 383, 173, 146], [ 71, 87, 498, 839], [ 20, 832, 759, 39], [461, 780, 518, 20], [118, 639, 26, 306], [264, 414, 258, 361], [ 18, 568, 439, 50], [612, 616, 332, 66]]...))>, 'class_idxs': [1, 2, 0, 0, 2, 0, 0, 0, 0, 0], 'keypoints': <PointsList(n=10)>, 'scores': [0.3595079 , 0.43703195, 0.6976312 , 0.06022547, 0.66676672, 0.67063787,0.21038256, 0.1289263 , 0.31542835, 0.36371077], 'segmentations': <SegmentationList(n=10)>, } >>> # xdoctest:+REQUIRES(--show) >>> import kwplot >>> kwplot.autompl() >>> dets.draw(setlim=True)
Example
>>> # Boxes position/shape within 0-1 space should be uniform. >>> # xdoctest: +REQUIRES(--show) >>> import kwplot >>> kwplot.autompl() >>> fig = kwplot.figure(fnum=1, doclf=True) >>> fig.gca().set_xlim(0, 128) >>> fig.gca().set_ylim(0, 128) >>> import kwimage >>> kwimage.Detections.random(num=10, segmentations=True).scale(128).draw()
- kwimage.structs.detections._dets_to_fcmaps(dets, bg_size, input_dims, bg_idx=0, pmin=0.6, pmax=1.0, soft=True, exclude=[])[source]¶
Construct semantic segmentation detection targets from annotations in dictionary format.
Rasterize detections.
- Parameters:
dets (kwimage.Detections)
bg_size (tuple) – size (W, H) to predict for backgrounds
input_dims (tuple) – window H, W
- Returns:
- with keys
size : 2D ndarray containing the W,H of the object dxdy : 2D ndarray containing the x,y offset of the object cidx : 2D ndarray containing the class index of the object
- Return type:
Example
>>> # xdoctest: +REQUIRES(module:ndsampler) >>> from kwimage.structs.detections import * # NOQA >>> from kwimage.structs.detections import _dets_to_fcmaps >>> import kwimage >>> import ndsampler >>> sampler = ndsampler.CocoSampler.demo('photos') >>> iminfo, anns = sampler.load_image_with_annots(1) >>> image = iminfo['imdata'] >>> input_dims = image.shape[0:2] >>> kp_classes = sampler.dset.keypoint_categories() >>> dets = kwimage.Detections.from_coco_annots( >>> anns, sampler.dset.dataset['categories'], >>> sampler.catgraph, kp_classes, shape=input_dims) >>> bg_size = [100, 100] >>> bg_idxs = sampler.catgraph.index('background') >>> fcn_target = _dets_to_fcmaps(dets, bg_size, input_dims, bg_idxs) >>> fcn_target.keys() >>> print('fcn_target: ' + ub.urepr(ub.map_vals(lambda x: x.shape, fcn_target), nl=1, sort=1)) fcn_target: { 'cidx': (512, 512), 'class_probs': (10, 512, 512), 'dxdy': (2, 512, 512), 'kpts': (2, 7, 512, 512), 'kpts_ignore': (7, 512, 512), 'size': (2, 512, 512), } >>> # xdoctest: +REQUIRES(--show) >>> import kwplot >>> kwplot.autompl() >>> size_mask = fcn_target['size'] >>> dxdy_mask = fcn_target['dxdy'] >>> cidx_mask = fcn_target['cidx'] >>> kpts_mask = fcn_target['kpts'] >>> def _vizmask(dxdy_mask): >>> dx, dy = dxdy_mask >>> mag = np.sqrt(dx ** 2 + dy ** 2) >>> mag /= (mag.max() + 1e-9) >>> mask = (cidx_mask != 0).astype(np.float32) >>> angle = np.arctan2(dy, dx) >>> orimask = kwplot.make_orimask(angle, mask, alpha=mag) >>> vecmask = kwplot.make_vector_field( >>> dx, dy, stride=4, scale=0.1, thickness=1, tipLength=.2, >>> line_type=16) >>> return [vecmask, orimask] >>> vecmask, orimask = _vizmask(dxdy_mask) >>> raster = kwimage.overlay_alpha_layers( >>> [vecmask, orimask, image], keepalpha=False) >>> raster = dets.draw_on((raster * 255).astype(np.uint8), >>> labels=True, alpha=None) >>> kwplot.imshow(raster) >>> kwplot.show_if_requested()
raster = (kwimage.overlay_alpha_layers(_vizmask(kpts_mask[:, 5]) + [image], keepalpha=False) * 255).astype(np.uint8) kwplot.imshow(raster, pnum=(1, 3, 2), fnum=1) raster = (kwimage.overlay_alpha_layers(_vizmask(kpts_mask[:, 6]) + [image], keepalpha=False) * 255).astype(np.uint8) kwplot.imshow(raster, pnum=(1, 3, 3), fnum=1) raster = (kwimage.overlay_alpha_layers(_vizmask(dxdy_mask) + [image], keepalpha=False) * 255).astype(np.uint8) raster = dets.draw_on(raster, labels=True, alpha=None) kwplot.imshow(raster, pnum=(1, 3, 1), fnum=1) raster = kwimage.overlay_alpha_layers(
[vecmask, orimask, image], keepalpha=False)
- raster = dets.draw_on((raster * 255).astype(np.uint8),
labels=True, alpha=None)
kwplot.imshow(raster) kwplot.show_if_requested()